to the Availability of RIKEN ENU-based gene-driven mutagenesis system (RGDMS) with the target-gene list and available mutant lines.
ENU-based gene-driven mutagenesis: NEXT-GENERATION GENE-TARGETING. [To Japanese Page]
Hot Topics: The RIKEN gene-driven mutagenesis system (RGDMS) is open to public within our capacity at the RIKEN BioResource Center. Established mutant lines are also available to research community. Click here or inquiry to;
[Please note that ENU mutant mice are non LMOs (non living modified organisms).]
WHAT'S NEW!
- [2010.4.12] The list of the target gene is updated. The list of available mutant lines is also updated. The application form and the PCR design manual are slightly modified. Accordingly, it becomes unnecessary to design multiplex PCR at all. Japanese version of our WEB site is now available.
- [2010.4.1] One hundred and four mutant strains established by gene-driven approach are now open and available.
- [2009.10.1] The discovered mutant mouse strains are now distributed by the BRC Experimental Animal Division. Accordingly, the costs for the rederivation and delivery are reduced.
- [2009.5.7] By trapping exon sequences of G1 mouse genomic DNAs and re-sequencing the trapped exon fragments, we now start feasibility study of making whole catalog of ENU-induced mutations in the exome of RIKEN Mutant Mouse Library.
- [2008.12.5] The list of the target genes is updated. Thirty five mutant lines carrying ENU-induced base-substitutions in the "long conserved noncoding sequences (LCNS)" are now available based on request. Japanese version of our WEB site is partly open.
- [2008.08.26] The WEB site is renewed under the Mutagenesis and Genomics Team at the RIKEN BioResource Center. The forms for the application to use the RIKEN ENU-based gene-driven mutagenesis system are slightly modified to reflect the affiliation change and others but the concept, system, how to use, and flow of the use are basically the same. The list of the target genes is also updated.
CONTENTS
1. BACKGROUND
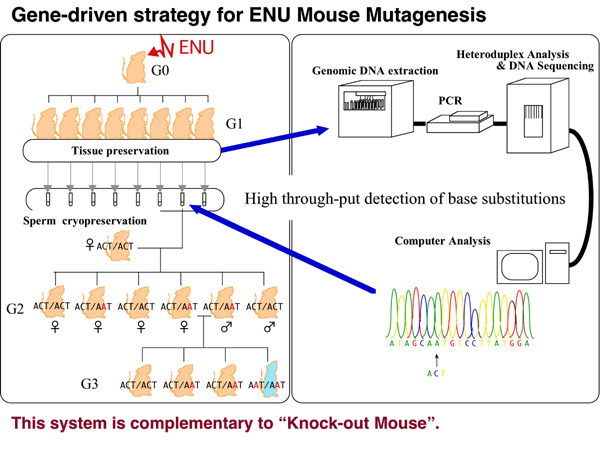
Functional Genomics Research Group at RIKEN Genomic Sciences Center archived ~10,000 G1 mouse sperm during the process of a large-scale ENU mouse mutagenesis during 2000 - 2008. The Population and Quantitative Genomics Team at RIKEN Genomic Sciences Center also constucted genomic DNA archive from the same set of G1 mice and developed a sequence-based screening system to conduct the ENU-based gene-driven mutagenesis. We call these dual archives "RIKEN mutant mouse libary".
The RIKEN Genomic Sciences Center closed March 2008 and the RIKEN mutant mouse library was transferred to RIKEN BioResource Center. To continue the Population and Quantitative Genomics Team's effort, a new team, Mutagenesis and Genomics Team was newly organized at the RIKEN BioResource Center as of April 2008. Now the Mutagenesis and Genomics Team is in charge of the ENU-based gene-driven mouse mutagenesis.
2. SYSTEM and STRATEGY
The Mutagenesis and Genomics Team utilizes the frozen sperm G1 mouse resource to conduct the ENU-based gene-driven mutagenesis. In addition to the sperm cryopreservation by the Mouse Functional Genomics Research Teams, we also keep several organs from each G1 male in order to archive genomic DNA from all the G1 males. By utilizing these genetic resources, feasibilities for gene-driven mutagenesis have been investigated. We now archived ~10,000 G1 male sperm and most of their genomic DNAs. We also established a mutation discovery system by using a temperature gradient capillary electrophoresis (TGCE) as a primary mutation screening system [1].
In addition, the Mutagenesis and Genomics Team is trying to accerelate the pace of the mutation discovery by using, for instance, TILLING/Cel1 digestion method, High Resolution Melt (HRM) methods, etc. Furthermore, the next-generation re-sequencing method is under investigation in order to thouroughly identify ENU-induced mutations in genomewide. At present, we are trying to enrich a part of or whole mouse exome and re-sequence by using one of the next-generation sequencers, for instance, Illumina, SOLiD and Helicos, which are currently available at the RIKEN Omics Research Center.
3. Publiations related to the RIKEN ENU-based gene-driven mutagenesis.
Review.
- Gondo, Y. (2008) Trends in Large-scale Mouse Mutagenesis: from Genetics to Functional Genomics. Nature Reviews Genetics 9: 803-810. (doi.org/10.1038/nrg2431)
- Gondo, Y., Fukumura, R., Murata, T., Makino, S. (2009) Next-generation gene targeting in the mouse for functional genomics. BMB Rep. 42(6): 315-323.
Mutation detection/rate/spectrum.
- Sakuraba, Y. et al. (2005) Molecular characterization of ENU mouse mutagenesis and archives. Biochem. Biophys. Res. Commun. 336: 609-619. (doi:10.1016/j.bbrc.2005.08.134)
- Takahasi, K.R. et al. (2007) Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Molecular Biology 8: 52. (doi:10.1186/1471-2199-8-52)
Targeted mutagenesis and established mutant lines.
- Masuya, H. et al. (2007) A series of ENU-induced single-base substitutions in a long-range cis-element altering Sonic hedgehog expression in the developing mouse limb bud. Genomics 89: 207-214. (doi:10.1016/j.ygeno.2006.09.005)
- Clapcote, S.J. et al. (2007) Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 54: 387-402. (doi:10.1016/j.neuron.2007.04.015)
- Erickson, R.P. et al. (2007) An N-ethyl-N-nitrosourea-induced mutation in N-acetyltransferase 1 in mice. Biochem. Biophys. Res. Commun 370: 285-288. (doi:10.1016/j.bbrc.2008.03.085)
- Sakuraba, Y. et al. (2008) Identification and characterization of new long conserved noncoding sequences in vertebrates. Mamm. Genome 19: 703-712. (doi:10.1007/s00335-008-9152-7).
- Labrie V. et al. (2009) Serine racemase is associated with schizophrenia susceptibility in humans and in a mouse model. Hum Mol Genet. 18(17): 3227-3243. (doi:10.1093/hmg/ddp261)
To Top
![]()

since 2008.8.26