to the RIKEN BioResource Center (BRC)
to the Mutagenesis and Genomics Team Homepage
to the ENU-based Gene-driven mutagenesis: NEXT-GENERATION GENE-TARGETING
to the Availability of RIKEN ENU-based gene-driven mutagenesis system (RGDMS) with the target gene list
Cf. This page was originally prepared by the Population and Qunatitative Genomics Team at RIKEN Genomic Sciences Center as;
"Bioinformatics for the Large-Scale Mouse Mutagenesis Project (Ver1.02) " by Y. Gondo.
New Information Biology: Population and Quantitative Genomics
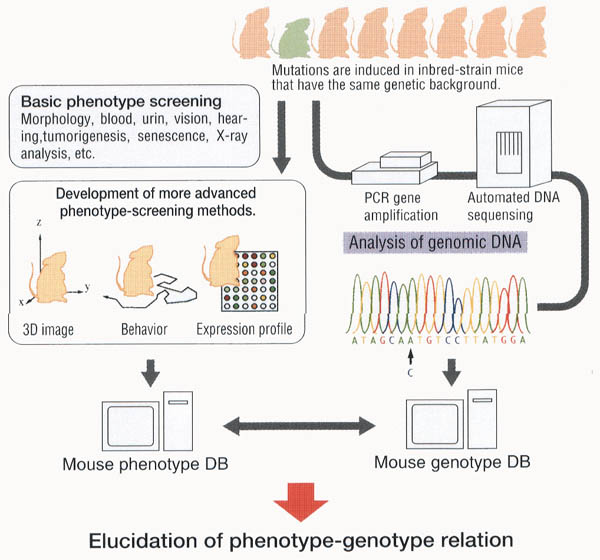
ENU-induced mutant mice carry single-base substitutions in their genomic DNA. ENU induces many single-base substitutions; therefore, the mutant mice have the causative point mutation for the mutant phenotype as well as many other silent mutations in the genome. In this sense, ENU-induced mutant mice are very similar to the situation of single nucleotide polymorphisms (SNPs) and will be a good animal model for the human SNP project. By using population and quantitative genetics, it is feasible to associate SNPs to genetic traits of diseases and individual's physiological conditions. However, it seems extremely difficult to conclude which SNP(s) is responsible for and causative of the trait or disease in humans. The animal model will provide a good experimental tool to investigate each SNP in terms of the biological function. In large-scale mouse mutagenesis, biological functions are compiled in a bulk phenotype database. At the same time, induced "SNPs" are sorted in the genotype database as described below. We refer to this genome-wide genetic approach with statistics and biometrics as population and quantitative genomics.

Mutations are first detected as a phenotype(s) different from that of wild-type mice in the large-scale mouse mutagenesis project. Then, the phenotype leads to identification and cloning of the causative gene(s) and SNP(s). Based on the phenotype, genetic mapping is conducted first to locate the chromosomal position of the mutation.Then, positional cloning identifies the gene and complete DNA sequencing determines the causative mutations or artificial SNP(s). In this approach, the majority of mutations will be dominant phenotypes and the pace of genotype database construction will be slow.
We preserve sperm samples from all the G1 males independently in liquid nitrogen tanks. We also isolate the genomic DNA from each G1 mouse. These archives of frozen sperm and genomic DNA consist of Mutant Mouse Library. From each G1's genomic DNA, target genes are amplified by PCR and mutations are identified using a heteroduplex detection system and DNA sequencing analysis. After finding mutations,candidate mutations are examined in vivo by retrieving the sperm from the liquid nitrogen and producing the viable mouse. In this sequence-based gene-driven approach, recessive phenotypes are easily examined because the mutation to be made for homozygotes is determined beforehand.
Data mining
From the phenotype database and genotype database, mutant candidates must be extracted effectively. At the same time, false positives must be carefully eliminated. The expected mutants in silico will be examined in vivo and vise versa. The genotypes are currently determined at the DNA sequence level although it is still laborious and expensive for genome-wide determination. On the other hand, phenotypes are a part of the whole biology and physiology and their measurement is occasionally very subjective. Therefore, it is necessary to establish the biometrics of the phenotype assessment so that each parameter is reproducible and reliable. By integrating these fundamental databases with population and quantitative genomics, a knowledge-based database for data mining becomes plausible. For these processes, new data-mining tools combined with biological and genetic tools are essential. The comparative genomics between human and mouse is also another key in effectively utilizing this animal model system to elucidate the mechanism of human diseases and to apply the outcomes to the medical and pharmaceutical field. It requires a high throughput system that is able to manipulate the huge complicated and diversified database.
Infrastructures for the Operation of the Database and Project
Bar-coding
Every week, ~100 G1 mice have been produced and all the males, whose sperm are stored in liquid nitrogen tanks, are maintained up to 78 weeks. Therefore, approximately 10,000 G1 mice are present in our facility at any given time. G1 parental mice, G2 and G3 mice and other necessary supporting mice (e.g., the mutation mapping scheme requires a number of mice) are also maintained in the specific-pathogen free (SPF) animal facility. Our facility requires a 50,000 mouse-cage capacity with a limited number of animal caretakers. A bar-coding system to identify each mouse is the key for efficient daily husbandry.
Local area network (LAN)
The mice must be protected from fatal infection by pathogens. Material transfer is, therefore, very restricted in order to maintain the SPF grade. Data transfer also requires no or minimal writing and copying errors. To achieve these conditions, all data compiled by the "input-only-once" manner at the original site directly into the computer and transferred into the ORACLE database simultaneously. In the mouse facility area, appropriate numbers of client computer equipments for data input is installed wherever necessary. LAN configuration and maintenance are thus extremely critical, since we minimized the use of pens and paper. Even one minute of LAN interruption could cause serious problems for these works.
Biometric devices and phenometrics
Various biometric devices are also located at many places for phenotype screening. At present, such equipments are usually controlled by a computer; however, in many cases the controlling computer is a stand-alone type and not connected to our LAN. To transport data from such isolated computers effectively, we construct a data-transfer protocol for each biometric device when necessary. Occasionally, the biometric device itself has to be modified, or even newly developed to achieve our purpose. Such renovation of biometrics, we call"phenometrics" should contribute a development of new diagnostic systems for humans.
Management tools
Daily activities and tasks are highly diversified at many different levels and categories in large-scale mouse mutagenesis. Animal caretakers, technical staff, researchers and other personnel must communicate with each other to coordinate the work. To organize daily, weekly and long-term plans in this complicated project, a "schedule planner" is necessary. An expert system for laboratory management and planning is ideal for this purpose. At the same time, control and maintenance of the facility are indispensable. Supplies of electric power, water, gas,etc. must be reliable and backup systems are periodically checked. The temperature, humidity, security locks and SPF grade are also monitored continuously and the logs are automatically transferred to the database server through LAN. If any emergency or problems should arise, the system must be able to trigger the alarm and safety systems including notification to respective staffs and offices.
Genetics
In order to produce ~100 G1 mice every week, mouse husbandry must be well planned a long time before hand, because each individual G1 goes through various examinations for 18 months. When the inheritance test is conducted for late-onset phenotypes, another 18 months are usually necessary. It also takes about one year from the ENU injection into G0 males to obtain G3 offspring. For mapping and molecular-cloning of identified mutations, additional time will be required. Mutant candidates must be carefully examined to eliminate false-positives and -negatives as minimum as possible at the very early stage of screening. Otherwise, manpower, space, and budgets would soon greatly exceed the capacity, or the candidates might all turn out to be false after a long screening and examination period. Key genetic factors are: 1) selection of inbred strain(s) to be used for ENU injection, for G1 production and G2 production, and for mapping, etc., and 2) effective inheritance tests with respect to environmental effects, degree of dominance, penetrance, heterosis, viability, sterility, meiotic drive, segregation distortion, genetic background, genetic drift and polygenic effects. Obviously the establishment of negative control data is essential for the entire project.
Ref. Y. GONDO (2001) "Bioinformatics for the Large-Scale Mouse Mutagenesis Project.", pp.763-767, in Knowledge-Based Intelligent Information Engineering Systems & Allied Technologies KES'2001 (Eds. by N. Baba, L. C. Jain and R. J. Howlett) IOS Press, Tokyo.
to the ENU-based Gene-driven mutagenesis: NEXT-GENERATION GENE-TARGETING
to the Availability of RIKEN ENU-based gene-driven mutagenesis system (RGDMS) with the target gene list
to the Mutagenesis and Genomics Team Homepage